Previous Issues Volume 8, Issue 1 - 2024
The Sea Star Anti-HRP Protein (IPA: Production in a CHO Protocol, Its Specificity in ELISA. Presence of CDR1 and CDR2 Regions. Aspects of Invertebrate Primitive Antibody
Michel Leclerc*
Immunology of Invertebrates, Orléans University, France
*Corresponding author: Michel Leclerc, Immunology of Invertebrates, Orléans University, France, Tel: 0238410209, Email: [email protected]
Received Date: May 03, 2024
Published Date: May 29, 2024
Citation: Leclerc M. (2023). The Sea Star Anti-HRP Protein (IPA: Production in a CHO Protocol, Its Specificity in ELISA. Presence of CDR1 and CDR2 Regions. Aspects of Invertebrate Primitive Antibody. Mathews J Immunol Allergy. 8(1):26.
Copyrights: Leclerc M. © (2024).
ABSTRACT
The sea star anti-HRP, we called, at the beginning: ALP (Antibody-like protein) reveals more and more, with the time, a quality of Invertebrate Primitive Antibody (IPA). We recall the existence of sea star lymphocytes, humoral specific adaptative immunity (anti-HRP protein or anti Horse-radish peroxydase), genomic data.In the precedent report we establish the production of this «Young Protein », its specificity anti-HRP by Elisa. At last we demonstrate the existence of CDR1 and CDR2 regions sharing strong similarities with the mammal: Pongo abelii.
Keywords: Invertebrates, CHO Protocol, Elisa, CDR1, CDR2, IPA.
INTRODUCTION
10 years ago, we tried to clone, for the first time, the Asterias rubens sea star IGKappa gene by the use and the help of E.coli as amplificator [1]. It allowed, in a second time, to verify that the Young Protein, or anti-HRP Protein recognizes the HRP antigen [1].
But, this verification of the affinity between the IPA (Invertebrate Primitive Antibody) and the antigen, seemed unclear at that time, for many of us.
Thus, we decided to operate a new cloning [2] of the IGKappa gene with new parameters and new affinity tests. This second one did not allow to obtain the protein of interest or « Young Protein ».
A third assay was attempted
It used a CHO method, as described in various experiments [3] and Elisa to verify the specificity of our « antibody » against the HRP.Otherwise, from the IMGT V domain we have researched the Complementary Determining region in the AA sequence of young protein.
MATERIALS AND METHODS
Construction
The nucleotide sequence of IPA was designed with optimized codon from Cricetulus griseus and with an histidine tag and a Ctag. Then it was synthesized by GeneArt in pcDNA3.4 plasmid (Thermo Fisher Scientific®). The plasmid obteined was transformed in DH5-α chemically competent bacteria. One clone was selectioned, amplified and purified by QIAGEN Plasmid Maxi Kit.
Production
Protein was produced by transient transfection of ExpiCHO cells according to the manufacturer’s instructions with max titer protocol (Thermo Fisher Scientific®, A29133). After eight day of production, cell viability was measured by using CytoSMART (Corning) and cells were centrifuged at 10000g for 20 min. Clarified supernatant was passed over a 0.22-µm filter and protein was purified with an Akta purifier (GE Healthcare Europe GmbH).
Purification
Purification was performed by CaptureSelect™ C-tagXL MiniChrom Column (ThermoFisher Scientific®, 5943072005). Briefly, column was stabilized with equilibration buffer (20mM Tris, ph 7.4), sample was passed through column, washed with 1,8 column volume of equilibration buffer and then 4 column volume of washing buffer (20mM Tirs, 1M NaCl, ph 7.4). This buffer passed until UV was stable. Protein was eluted with 0.1 M citrate gradiant, pH 6 to pH3 and desalted through the HiPrep 26/10 desalting column (Cytiva, 17508701) in PBS buffer (2.7 mM KCl, 0.14 M NaCl, 1.5 mM KH2PO4, 8 mM Na2HPO4, pH 7.4). The concentration was determined with an UV detector at 280 nm. Protein molecular mass and molar extinction coefficient data were generated by using the Protparam tool (http://web.expasy.org/protparam/).
Size-exclusion chromatography (SEC) was performed on a Superdex 200 increase 10/300 GL, column (Cytiva, 28990944). The column was used at a rate of 0.4 mL/min with a UV detector at 280 nm. After loading, the protein was eluted in PBS with 1,2 column volume.
Analyse
SDS-PAGE
The integrity of protein was assessed by SDS-PAGE on 10% polyacrylamide gel under reducing or non-reducing conditions and Coomassie blue staining. Samples were all loaded pur and migrated at 200 V, 400mA, 35 min. Color Prestained Protein Standard, Broad range (New England BioLabs®, P7719S) was used.
ELISA
Affinity of protein was evaluated by ELISA (enzyme linked immunosorbent assay). Protein was coated in 96-well plate after serial dilution in PBS and incubated overnight at +4°C. After saturation in PBS-BSA (Bovin serum albumin) 3% during two hours at 37°C and washing with PBS-Tween 0.05%, a protein-L-peroxydase (ThermoScientific, Pierce®, 32420) or a Streptavidin-peroxydase (ThermoScientific, Pierce™, 21130) was added at respectively 1.25 μg.mL-1 or 100 ng.mL-1 and incubated for one hour at 37°C. Enzymatic reactions were started by added of 100µL of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Serva Electrophoresis, 37068) and stopped with 50 μL of 1 M H2SO4. The absorbance was measured at 450 nm, using an absorbance microplate reader (Bio-Tek® instruments).
DETERMINATION OF DETERMINING REGION (CDR1, CDR2, CDR3))
Ii was made according the method of Ehrermann and Lefranc [4,5] from the IMGT V domain.
RESULTS
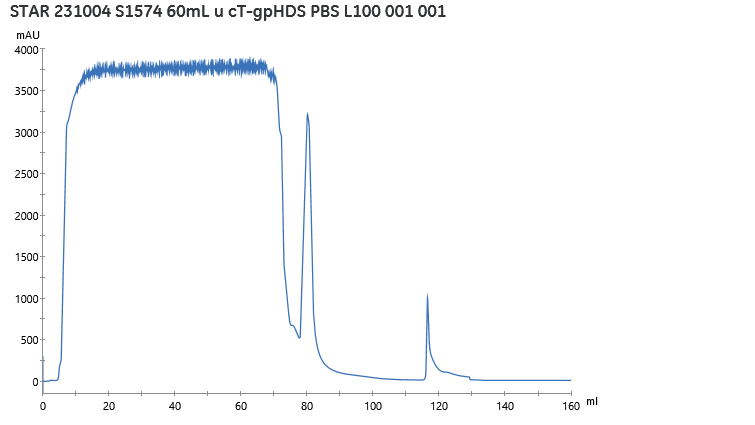
This protein was produced in ExpiCHO and purified by affinity for Ctag. The purification profile obtained is shown in Figure 1. From 0 to 65mL, supernatant of cell culture was passed through the column. The column was equilibrated from 65 to 74 mL and washed from 74 to 94 mL. The elution gradient started at 110 mL and protein was detected from 115 mL to 130 mL. During the column wash, a peak around 3000mAU was observed due to non-specific dropout. The elution started at 36% of pH 3, around pH 4.70. The peak elution starts with a steep slope but descends slowly, due to a possible impurity of protein or different forms thereof.
Figure 1. Profil of protein purification, observation of UV at 280nm during sample aplication, equilibration, washing and elution phase.
The elution phase with protein of interest was desalted through the desalting column and obtained in PBS. At the end of this purification process, 4mL of protein at 283.6 mg/mL and 10 mL of protein at 134.5 mg/mL were obtained, giving a total of 2.48 mg of protein of interest. This quantity corresponds to a production yield of 44.8 mg/L of cell culture.
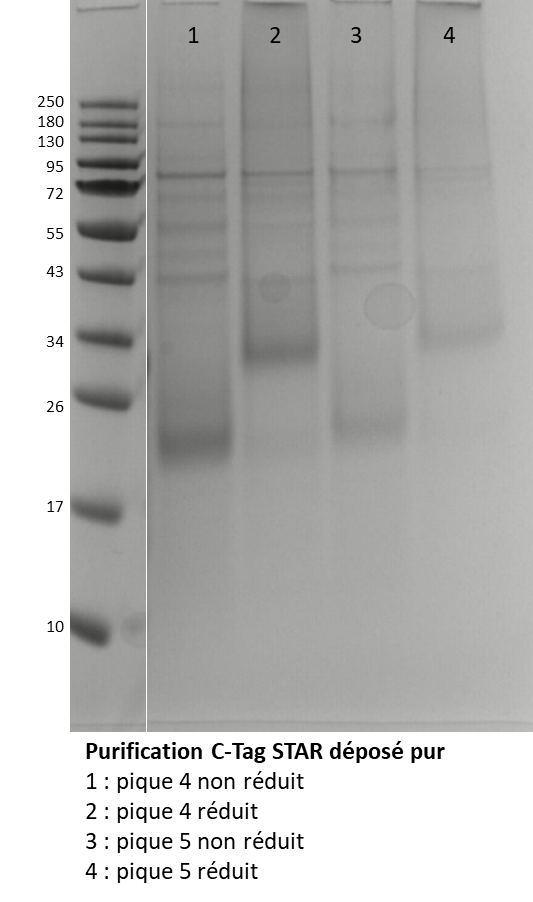
Two fractions of protein were obtained, representing two parts of the peak elution. They were analysed by SDS-PAGE to observe their integrity.
Figure 2. SDS-PAGE of protein purified after desalting. Fraction at 283.6mg/mL in unreduced (1) and reduced (2) condition and fraction at 134.5 mg/mL in unreduced (3) and reduced (4) condition.
The gel SDS-PAGE, in Figure 2, shown two fractions were similar, the protein of interest was shown arround 20 KDa in no reduced condition et arround 34 KDa in reduced condition. However, the presence of many other bands indicates the presence of impurities. Size Exclusion Chromatography (SEC) was performed to try to remove these impurities after injection of 2 mL fraction at 283.6 mg/mL.
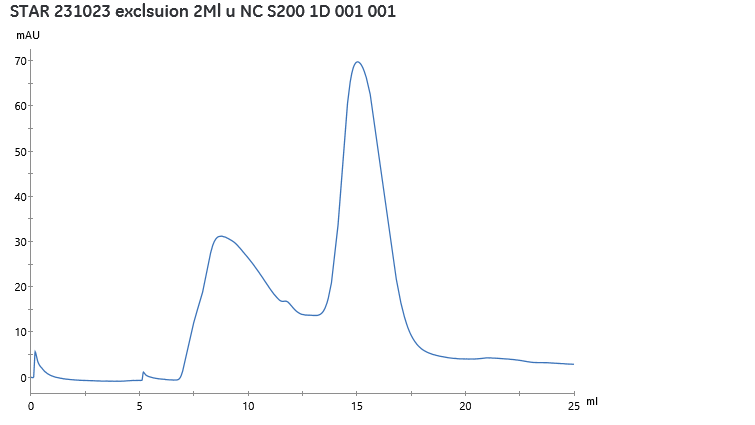
Figure 3. Size-exclusion chromatography realised after injection of 2mL of fraction at 283.6 mg/mL, obseration of UV at 280nm during elution of protein through S200 increase in PBS.
Figure 3 revealed the presence of two forms of the protein. The monomeric form, eluted at 15mL, represents 53% of the observed peaks. After pooling of the eluted monomeric peak, the fraction was 2 mL containing 24.85 mg/mL of protein of interest.
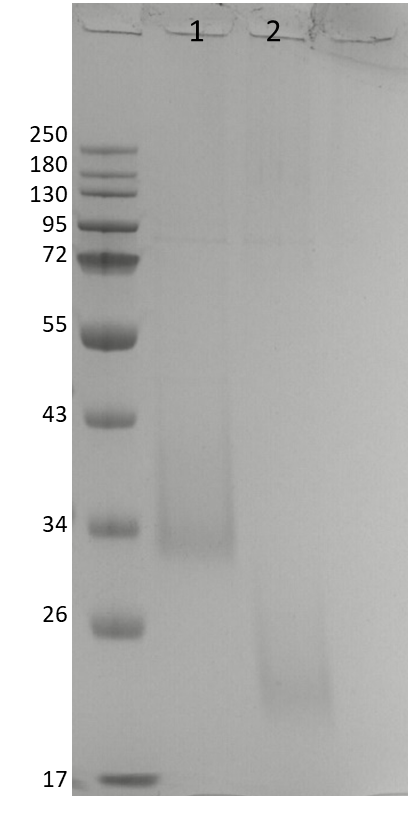
This purified fraction was then analysed by SDS-PAGE to compare purity after SEC. The result, shown in Figure 4, was a protein without impurities at 34 KDa in the reduced state or at around 20 KDa in the unreduced state.
Figure 4. SDS-PAGE of protein purified after SEC. Fraction at 24.85 mg/mL in reduced (1) and unreduced (2) condition.
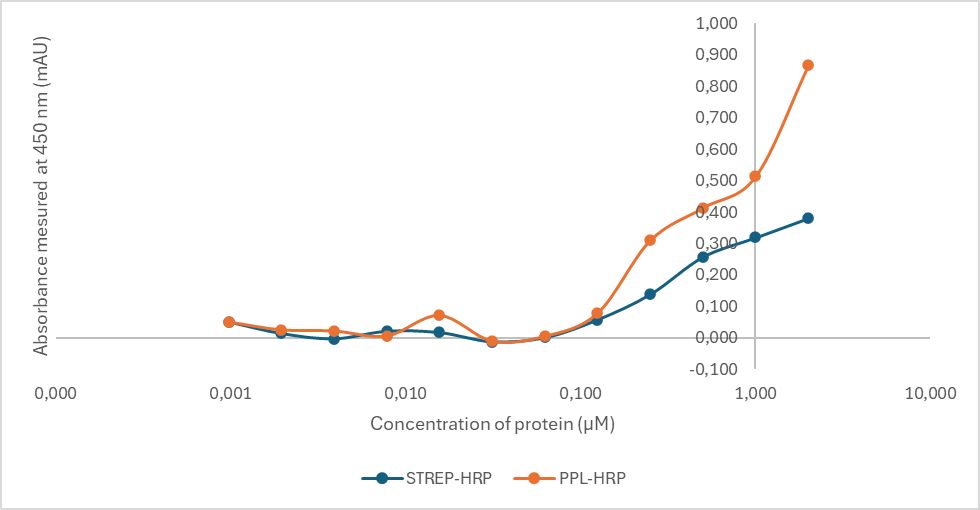
The affinity of the protein for peroxidase was validated by ELISA. The fraction of monomeric forms was added to the wells after serial dilution to 2 at 0.001 µM in PBS. Figure 5, shows the binding of protein L (ppl) and streptavidine (Strep) conjugated to peroxidase (HRP) up to a concentration of 0.250 µg/mL of protein of interest.
Figure 5. ELISA proteine of interest coupled with proteine L (ppl) or streptavidine (Strep) conjugated to HRP.
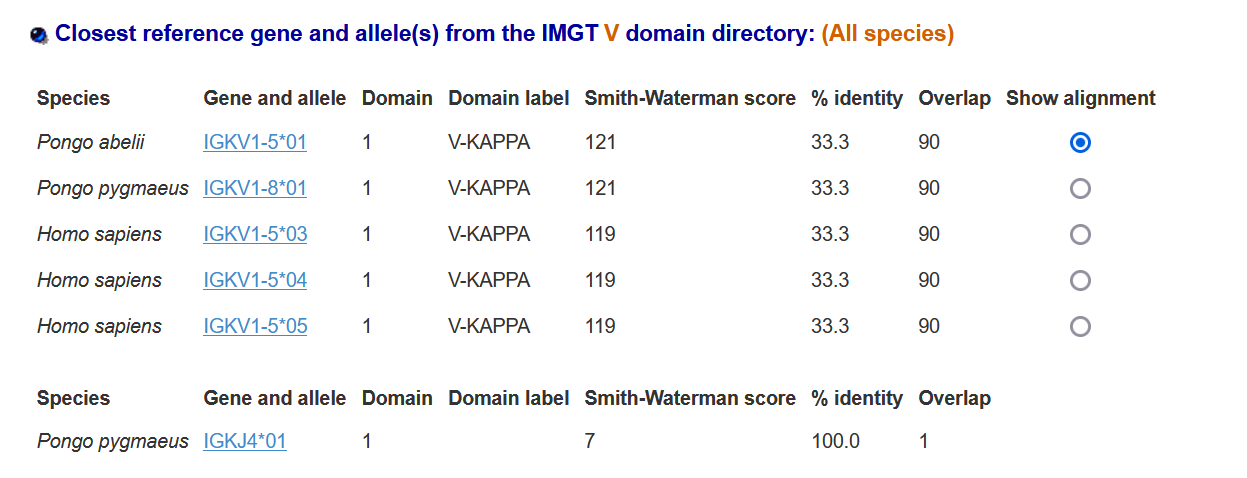
Results obtained with IMGT V domain, concerning CDR are repertoried in following tables:
IMGT/DomainGapAlign version: 4.10.3 (2021-12-06): Table I
II) Table II: Alignments:
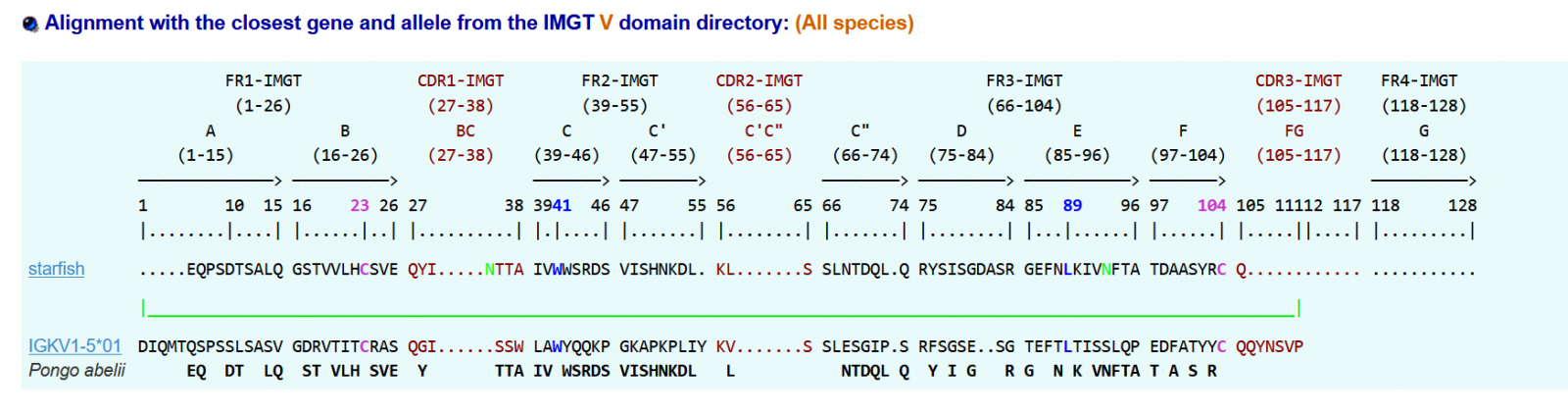
>seastar|IGKV1-5*01|33.3|||Pongo abelii
.....EQPSDTSALQGSTVVLHCSVEQYI.....NTTAIVWWSRDSVISHNKDL.KL.......SSLNTDQL.QRYSISGDASRGEFNLKIVNFTATDAASYRCQ..….
DISCUSSION AND CONCLUSION
The anti-HRP obtained purification° shows clearly
A good ratio was shown in CHO protocol as seen in purity and rate: the better we have, since we work in the sea star model. It’ll allow to serve as guide, in further experiments.
For the second time, the antigen HRP is recognized by the sea star antibody
The first time [1] was situated in 2014, after the isolation of sea star IGKappa gene(affinity with the enzyme HRP) and the second one, appears in 2024, with ELISA which is more specific in terms of specificity. 2 conjugates (L Protein and streptavidin associated respectively with HRP bind the antigen HRP).
At last Complementary Determining regions were discovered by the use of IMGT V domain
The conserved amino-acid positions 23 41 89 104 were found in the sea star sequence. Our molecule appears to have an IGAA sequence. It aligns with the mammal: Pongo
IGKV1-5 and the percentage of alignments was 33 %: a similarity to an IGKV gene when it comes to conserved amino-acids
CONCLUSION
We mean, you’ll understand (it’s not a problem of semantic but a question of evidences) we have to speak not of IG LIKE when compared to the sea star Anti-HRP protein but of a sort of Invertebrate Primitive Antibody:
TO PRESENT SEA STAR LYMPHOCYTES [6] It is a beginning of Invertebrate Primitive Antibody
TO PRESENT SEA STAR LYMPHOCYTES AND HUMORAL SPECIFIC RESPONSES: it’s better [7]
TO PRESENT SEA STAR LYMPHOCYTES, HUMORAL SPECIFIC RESPONSES AND GENOMIC DATA [8] SUCH INVERTEBRATE MHC GENES (Class I and Class II), Fab, Fc GENES: it’s better of better.
Moreover, CDR1 and CDR2 have been identified. At our knowledge, no heavy chain was found in our molecule but just light chain [8] (Kappa one). We have to compose with such a model of IPA. And CDR regions, in the future. Last observations made by the mean of modelization in 3D seem to show it exists also a CDR3 determining region.
ACKNOWLEDGMENTS
Dr N. Aubrey and his team for his SKILLFULL ASSISTANCE Faculty of Pharmacy Tours France.
REFERENCES
- Vincent N, Osteras M, Otten P, Leclerc M. (2014). A new gene in A. rubens: A sea star Ig kappa gene. Meta Gene. 2:320-322.
- Leclerc M. (2023). Other sea star Igkappa gene cloning assay in E. coli with new parameters. J Stem Cell Res The. 8(1):25-26.
- Polonelli L, Pontón J, Elguezabal N, Moragues MD, Casoli C, Pilotti E, et al. (2008). Antibody complementarity-determining regions (CDRs) can display differential antimicrobial, antiviral and antitumor activities. PLoS One. 3(6):e2371.
- Ehrenmann F, Kaas Q, Lefranc MP. (2010). IMGT/3Dstructure-DB and IMGT/DomainGapAlign: a database and a tool for immunoglobulins or antibodies, T cell receptors, MHC, IgSF and MhcSF. Nucleic Acids Res. 38(Database issue):D301-D307.
- Ehrenmann F, Lefranc MP. (2011). IMGT/DomainGapAlign: IMGT standardized analysis of amino acid sequences of variable, constant, and groove domains (IG, TR, MH, IgSF, MhSF). Cold Spring Harb Protoc. 2011(6):737-749.
- Leclerc M, Arneodo VJ, Legac E, Bajelan M, Vaugier GL. (1993). Identification of T-like and B-like lymphocyte subsets in sea star Asterias rubens by monoclonal antibodies to human leukocytes. Thymus. 21(3):133-139.
- Brillouet C, Leclerc M, Binaghi RA, Luquet G. (1984). Specific immune response in the sea star Asterias rubens: production of "antibody-like" factors. Cell Immunol. 84(1):138-144.
- Vincent N, Osteras M, Otten P, Leclerc M. (2014). A new gene in A. rubens: A sea star Ig kappa gene. Meta Gene. 2:320-322.