Previous Issues Volume 8, Issue 1 - 2024
Modelizations in 3D of the Anti-HRP Sea Star (or Starfish) Primitive Antibody (IPA): Their Relations with Mus Musculus Fab and In A General Way Mammal Antibodies
Michel Leclerc*
Immunology of Invertebrates, Orléans University, France
*Corresponding author: Michel Leclerc, Immunology of Invertebrates, Orléans University, France, Tel: 0238410209, Email: [email protected]
Received Date: May 11, 2024
Published Date: June 03, 2024
Citation: Leclerc M. (2023). Modelizations in 3D of the Anti-HRP Sea Star (or Starfish) Primitive Antibody (IPA): Their Relations with Mus Musculus Fab and In A General Way Mammal Antibodies. Mathews J Immunol Allergy. 8(1):27.
Copyrights: Leclerc M. © (2024).
ABSTRACT
Presentations of sea star primitive antibody 3D modelizations, according AlphaFold and Swiss Model predictions were performed in EMBL Grenoble A superposition of the 2 predictions lead us to envisage a true link with a « part » of Mus musculus IG antibody: the Fab one of Heavy Chain. But, at every time, in such presentations, it could be contested by those who don’t believe in the existence of the IPA (Invertebrate Primitive Antibody) for various reasons we don’t want enumerate.
Keywords: Invertebrates, CHO Protocol, Elisa, CDR1, CDR2, IPA.
INTRODUCTION
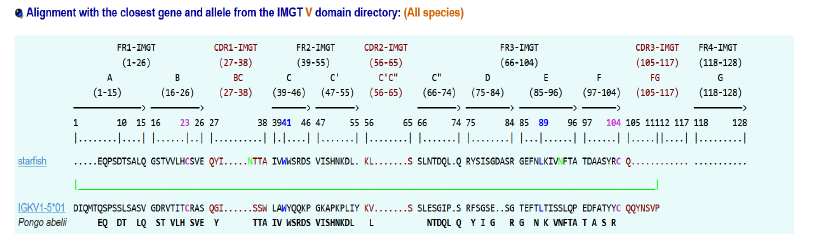
Recently, (2024) we have discovered CDR1, CDR2, CDR3? In sea star Immune system [1] with the help of Dr Sofia Kossida (IMGT Montpellier) [2,3]. We try in the present study to give modelizations of the sea star IGKappa protein [4] which recognizes specifically the antigen HRP. In the first part we recall the different data and characteristics of sea star IGKappa: i-e sequence and different alignments (aa) when compared to mammal sequences in aa. At last we attempt to find again, in the 3D modelization, the « regions »which may be compared to CDR1, CDR2, and CDR3.
MATERIALS AND METHODS
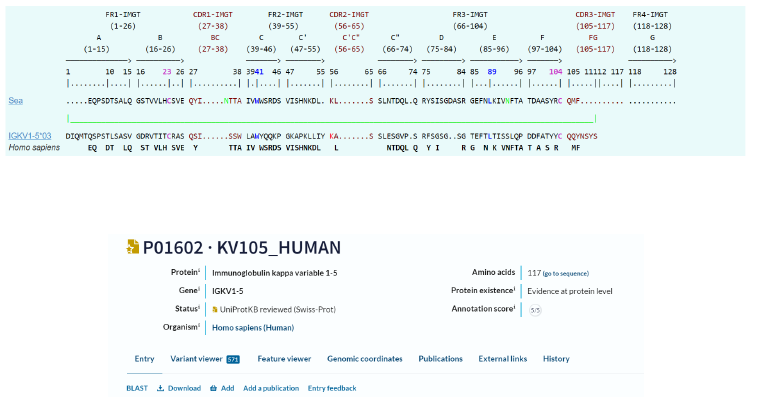
Alphafold and Swiss Model entities, which predict a protein’s 3D structure based on its primary amino acid sequence, were used. Recalling of IGKappa gene sequence and comparison of aa alignments between this last one and mammal ones are first given:
Inputs: IMGT/DomainGapAlign version: 4.10.3 (2021-12-06)
Sequence:
MRGNMASLWMFFFVVGITLQRSLAIYTFREQPSDTSALQGS
TVVLHCSVEQYINTTAIVWWSRDSVISHNKDLKLSSLNTDQ
LQRYSISGDASRGEFNLKIVNFTATDAASYRCQMFA
RESULTS
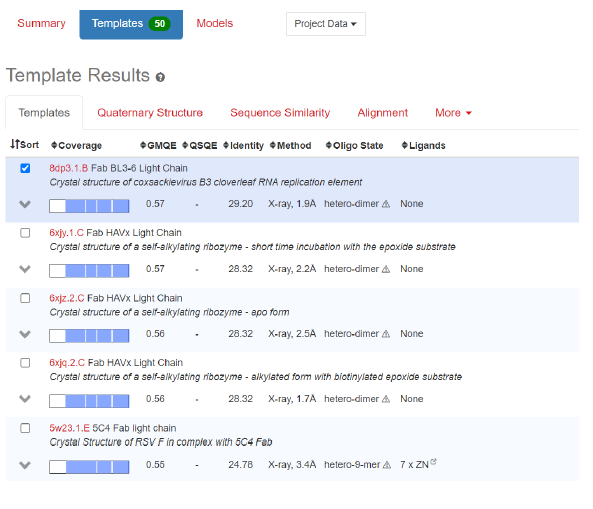
1. Comparisons with Fab Light chain(crystal): Swiss Model (https://swissmodel.expasy.org/)
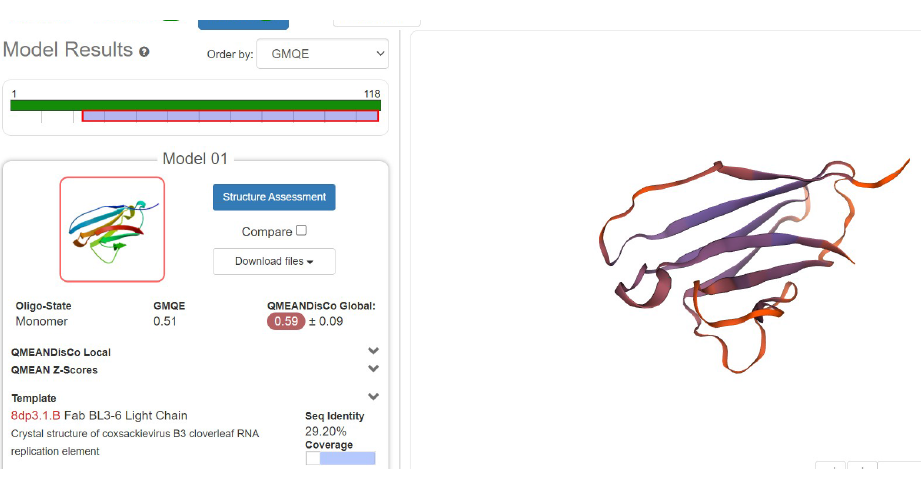
2. First Modelization of anti-HRP protein
3. AlphaFOLD
5 models predicted and for each 4 seeds are tested → 20 predictions
Superimposition done from residue 26
The RMSD is on average from 0.6 to 0.3 A
The only variability is at the level of α-helix
4. Parameters of the alphafold prediction
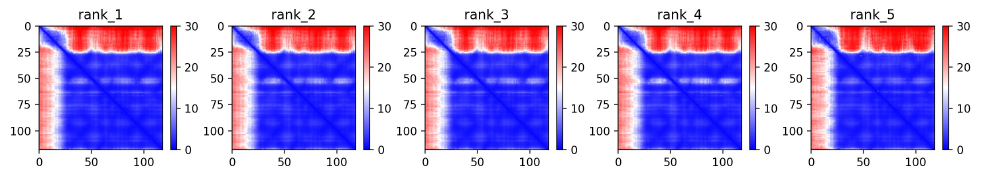
Predicted alignment error plot (PAE). The absolute error in A of the relative position betweenresidues
Local confidence of the model:
- Dark blue: large confidence
- Red: low confidence
0-25: red → low confidence (up to 30 A of error)
25-118: blue with some regions in white → more confident
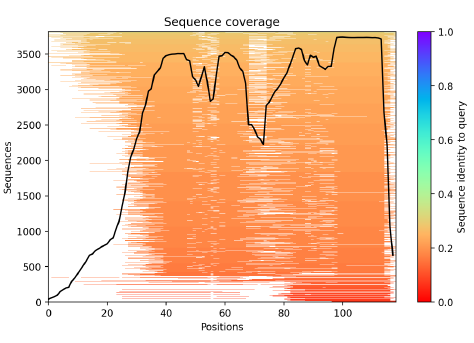
5. Multiple-Sequence Alignment (MSA)
The sequences used in the alignment ordered from largest identity (top) to the lowest identity (bottom). White parts are not covered. The black lines indicate the coverage of the sequence.
0-24 residues: not well covered by the sequence alignment → α-helix predicted
24- 118 residues: identity found in the range of 0.2-0.3, the pic is for the last 30 residues
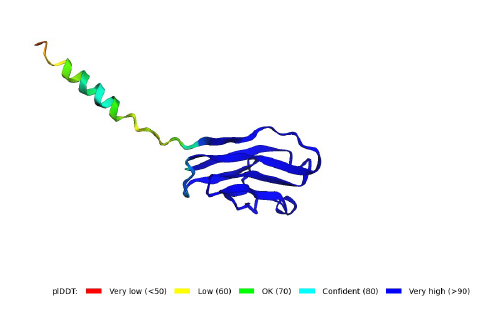
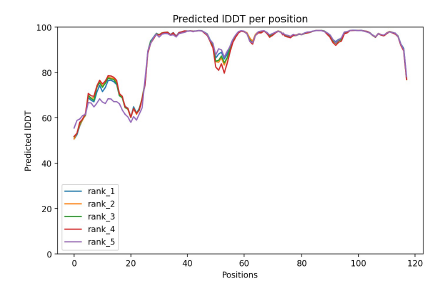
6. lDDT: Local Distance Difference Test
It evaluates local distance differences of all atoms in a model including validation of stereochemical plausibility.
plDDT > 90 modelled to high accuracy
plDDT between 70 to 90 modelled well
plDDT between 50 and 70 low confidence
0-20 residues have a plDDT between 80 and 50
→ this region is majoritary modelled with low confidence, with a pick at 80 which indicated that a portion is well modelled 20-118 residues have a lDDT higher than 90 however some regions (around position 50 and after 110) have a score between 90 and 70 → this regions is model with high accuracy with two regions well modelled.
7. SCALOP: Sequence-based antibody Canonical LOoP structure annotation
(https://opig.stats.ox.ac.uk/webapps/sabdab-sabpred/sabpred/scalop)
SCALOP is a sequence-based canonical form predictor for five of the six complementarity determining regions (H1, H2, L1, L2 and L3) on an antibody.
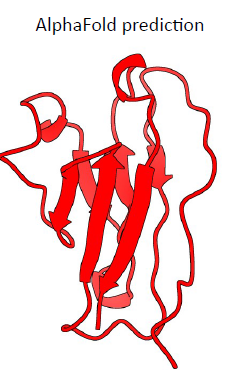
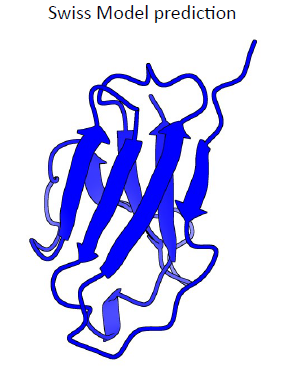
8. Primitive invertebrate antibody – final predictions
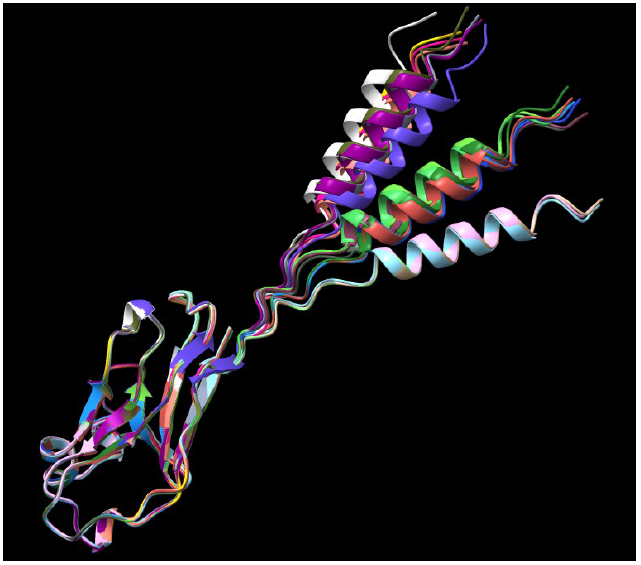
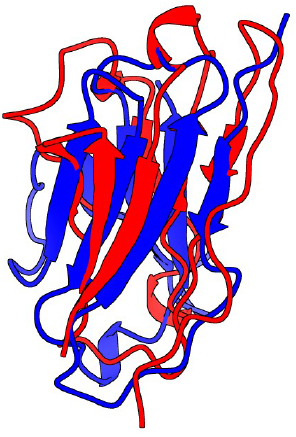
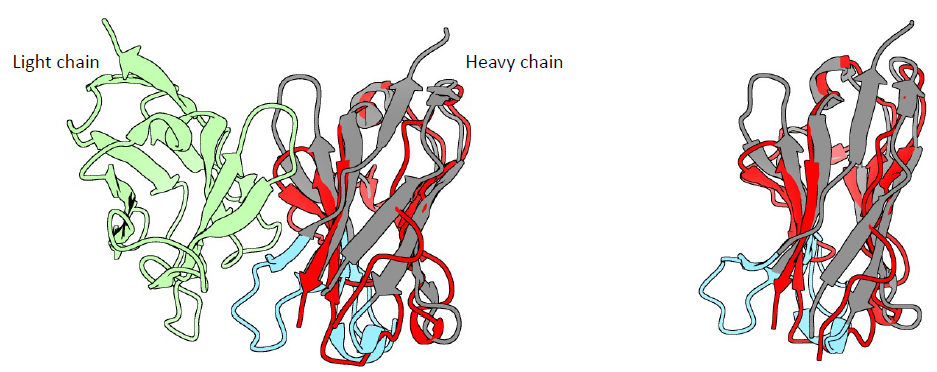
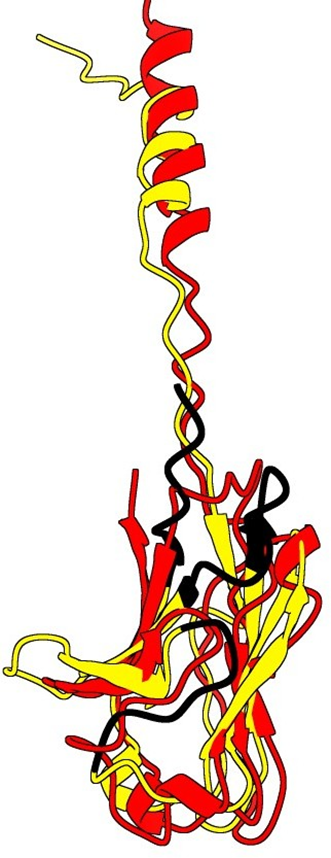
9. Superpose of the predictions
Red: Alpha Fold prediction
Blue: Swiss Model prediction
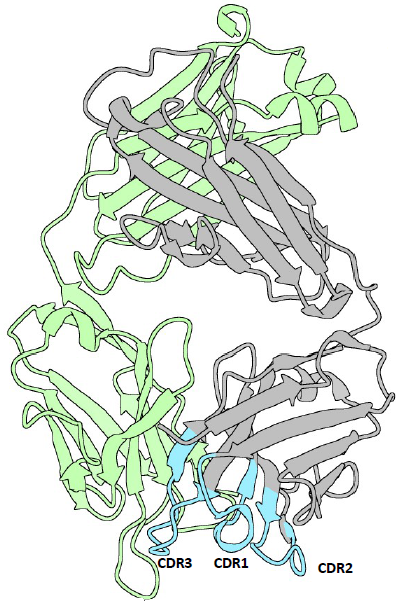
10. 4HDI : 3E5 IgG3 FAB from mus musculus
Green: light chain
Grey: heavy chain
Light blue: CDRs
CDR1: 26-35
CDR2: 50-58
CDR3: 99-109
https://www.rcsb.org/structure/4HDI
11. Superpose of AlphaFold prediction and X-ray structure of a Fab (4HDI)
The SSM superpose in Coot aligns the prediction with the heavy chain
12. IMGT/DomainGapAlign
13. AlphaFold prediction of IGKV1-5*03 Homo sapiens
CDR1: 46-55 residues
CDR2: 72-78 residues
CDR3: 111-117 residues
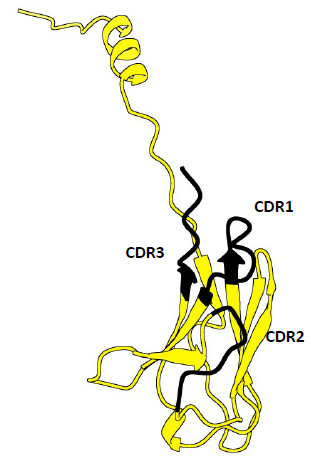
14. AlphaFold predictions superposed
Yellow: IGKV1-5*03 Homo sapiens
Red: Invertebrate Primitive Antibody from Starfish
Possible CDRs based on the superposition
CDR1: 48-58
CDR2: 69-73
CDR3: 115-118
DISCUSSION AND CONCLUSION
It seems clearly that our primitive antibody shares similarities with the Fab (4HDI) from the Heavy chain of Mus musculus in terms of amino-acids when we have a look to CDR1:26-35 residues. CDR2: 50-58 residues CDR3: 99-109 residues and in terms of morphology in 3D (Modelization), when compared always to the Fab ‘4HDI: 3E5 IGG3 Fab. No similarities were found with THE LIGHT CHAIN, except in the model 01, where it is shown slight similarities (Sequence identities of 29, 20 %) with Fab BL3-6 Light Chain: what is a weak percentage.
A comparison of our antibody with IGKV1-5 03 was also productive in terms of alignments for CDR1: 46-55 residues, CDR2:72-78 residues, CDR3: 111-117 residues. These last predictions were done with AlphaFold ones. This work presents the interest to name CDR3 in our research. It was not the case in our precedent work (1) our primitive antibody would present CDR1, CDR2, CDR3 (determining regions) as seen precedently and, more specially, similarities with Fab (4HDI) (by the superpose in Coot aligns the prediction with the Heavy Chain). Less with IGKV1-5 03 Homo sapiens. The main work is now to persuade that it consists in true observations+ and confirm the existence of the IPA (Invertebrate Primitive Antibody).
+We thanks greatly the Doctor J.A Marquez: EMBL (Grenoble) and his laboratory for their skillful and technical competences in this work in which, more and more people, dare to speak of Invertebrate Primitive Antibod.
REFERENCES
- Leclerc M. (2024). Determination of the CDR (CDR1, CDR2) Complementary-Determining Region Invertebrate Primitive Antibody from Sea Star. Mathews J Immunol Allergy. 8(1):25.
- Ehrenmann F, Kaas Q, Lefranc MP. (2010). IMGT/3Dstructure-DB and IMGT/DomainGapAlign: a database and a tool for immunoglobulins or antibodies, T cell receptors, MHC, IgSF and MhcSF. Nucleic Acids Res. 38(Database issue):D301-D307.
- Ehrenmann F, Lefranc MP. (2011). IMGT/DomainGapAlign: IMGT standardized analysis of amino acid sequences of variable, constant, and groove domains (IG, TR, MH, IgSF, MhSF). Cold Spring Harb Protoc. 2011(6):737-749.
- Vincent N, Osteras M, Otten P, Leclerc M. (2014). A new gene in A. rubens: A sea star Ig kappa gene. Meta Gene. 2:320-322.
.png)