Previous Issues Volume 8, Issue 10 - 2023
Antibiogram Profile of Bacteria of Public Health Importance Potentially Isolated From Domestic Dogs in Eleme Locality, Rivers State, Nigeria
Azuonwu O1,*, Clement Tamnyegia John1, Felix Barikuura Dimkpa2
1Department of Medical Laboratory Science, Rivers State University, Nkpolu-Oroworukwo, Port Harcourt, Nigeria
2Department of Medical Laboratory Science, Rivers State College of Health Science and Management Technology, Port Harcourt, Nigeria
*Corresponding Author: Azuonwu O, Rivers State University, Port Harcourt, Nkpolu–Oroworukwo, Port Harcourt, Rivers State, Nigeria; Email: [email protected]
Received Date: November 02, 2023
Publication Date: December 05, 2023
Citation: Azuonwu O, et al. (2023). Antibiogram Profile of Bacteria of Public Health Importance Potentially Isolated From Domestic Dogs in Eleme Locality, Rivers State, Nigeria. Mathews J Case Rep. 8(10):135.
Copyright: Azuonwu O, et al. © (2023)
ABSTRACT
Dogs are potential source of pathogenic bacteria of zoonotic importance across the world, especially in communities that lack adequate veterinary services. This study was aimed at investigating pathogens associated with domestic dogs and determining their susceptibility pattern to selected antibiotics. A total of fifty (50) samples were collected randomly from different locations within Eleme Local Government Area, Rivers State, Nigeria, from the buccal and nasal cavity of domestic dogs. The bacteriological analysis was determined using standard microbiological techniques of culture, isolation, identification, and antibiotic susceptibility testing. A total of six pathogens were isolated which include Bacillus spp., Candida albicans, Escherichia coli, Klebsiella spp., Proteus spp., and Staphylococcus aureus with prevalence of 2%, 8%, 21%, 20%, 7% and 10% respectively with E.coli and Klebsiella spp. being the most predominant. The study also shows that pathogens were susceptible to ciprofloxacin, gentamycin and amoxicillin with susceptibility rate of 62%, 57% and 53% respectively. However, Cefuroxime showed high resistant of 66%. Hence, this study is suggestive that oral and nasal droplets from dogs carry potential pathogens of public health concern, which may increase the burden of antibiotic-resistant strains pathogens in our weak health facilities and communities in general.
Keywords: Antibiogram, Antibiotics, Domestic Dogs, Nigeria, Public Health
INTRODUCTION
The past few decades have witnessed potential increase in pet animals including Canis lupus familiaris (Domestic dog) and due to their constant sharing of space and interaction with humans; it has consequently led to a surge in rate of infectious diseases globally. According to global pet ownership statistics, more than half the world population owns a pet. The American Medical Veterinary Association (AMVA), also reported that 57 percent of US households own at least 1 companion animal with about 150 million dogs [1-3]. Nonetheless, domestic dogs are reservoir of many pathogens of zoonotic significance which are transmissible through obvious means including saliva, aerosols, contaminated urine or faeces and direct contact with dogs [4]. In Africa and developing countries, about 20% of human morbidity and mortality are attributed to endemic zoonosis [5,6]. There are ecological changes which favour the development of some vectors of infection; urbanization which has led to increase contact with wildlife, hence have constitute a significant reservoir of zoonotic infectious diseases as well as other socio-cultural behaviour such as farming, hunting, and tourism which further encourage the emergence and re-emergence of zoonotic infectious diseases [7].
Nonetheless, different kinds of pathogens are associated with pet dogs and these could result to varieties of infectious diseases when transmitted to humans. They cause different kinds of illnesses in animals and humans, ranging from minor skin infections to serious diseases. Saliva of dogs are known to be potential source of an emerging strain Staphylococcus aureus called methicillin-resistant Staphylococcus aureus (MRSA) and Capnocytophaga canimorsus, an organism carried in the mouths of dogs, and it causes a very bad sepsis infection [8]. Vectors and vector-borne pathogens (VBPs) are also associated with pet animals. They are organisms that spread infections by conveying pathogens from one potential host to another causing lethal infection in domestic dogs and thus have strong consequence of zoonotic repercussion via animal to pet relationship. Most often these pathogens include Borrelia bugdorferi, Anaplasma phagocytophilium, and Dirofilaria immitis [9]. Dog is a reservoir of zoonotic infections. Most viral infections including rabies and norovirus, and bacterial infections including Leptospira, Brucella, Salmonella, Pasteurella, Staphylococcus intermedius and methicillin-resistant Staphylococcus aureus, fungal (e.g. dermatophytes), and parasites (e.g. Toxoplasma gondii) are common zoonotic infections transmitted to humans [4,10]. They are acquired through contact with animal saliva, urine, and other body fluids or secretions, ingestion of animal faecal material, inhalation of infectious droplets, and through bites, scratches and other direct contact with animals [11].
Infectious diseases have created strong public health implications and diverse biological nuisance to our environment. Notwithstanding, the sole aim of public health as the science and art of preventing disease, prolonging life and promoting human health through organized efforts and informed choices of society, organizations, public and private, communities and individuals [12,13] will be defied if adequate precautionary measures are not imbibe in day-to-day interaction with these pet animals including domestic dogs. Pet animal however, in spite of the worthy benefits to the society and homes, companion dogs remain a major threat to public health, with dogs harbouring several pathogens which are transmissible to humans. Therefore, with the increasing number of pet dogs in our society today, there is proportionate increase in contagion between dogs and people, together with a low level of hygienic conditions and there is also lack of sufficient veterinary attention and zoonotic disease awareness which compound the risk of transmission of zoonotic gastrointestinal parasites and other microorganisms to humans [14].
Zoonosis is an infectious disease transmitted from animals to human. However, diseases previously confined in certain part of the globe have appeared in new areas, hence it is imperative to recognize new diseases and to determine their causative agents. The most promising method of assessing new disease involve parallel and multidisciplinary approaches such as clinical recognition, epidemiology studies, standard laboratory assessment and laboratory testing [15,16]. The most common zoonotic diseases of dogs that fall in this category include dermatophytosis, campylobacter infection, leptospirosis, lyme disease, salmonellosis, roundworm, cryptosporidium infection, tapeworm, hookworm and rabies. Previous studies reported that some of these diseases are characterized by diarrhea as one of the most clinical common manifestations which can potentially lead to severe dehydration; hence zoonosis has been reported as one of the leading causes of death [17].
Notwithstanding, developing countries around the world have the highest global burden of some zoonotic diseases commonly encountered through bites from dogs. In Nigeria, few concerns are on diseases transmitted from domestic dogs to human. For example, rabies, a zoonotic disease encountered through dog bite has experienced ineffectiveness of eradication program. Reports have shown that exposure to rabies through the bite of dog account for 100% of confirmed cases in Nigeria [18,19]. Majority of other zoonotic diseases from domestic dogs are given less attention unlike other countries around the globe. However, there is paucity of information in regard to diseases that are transmitted from domestic dogs to human which made this study essential. Though few existing studies coupled with diverse conclusions have encouraged researchers to consider this direction of study particularly with the increase in demand of domestic dogs for hunting, security and as pets globally. Therefore, this study was aimed at isolating and characterizing bacteria pathogens associated with household pet dogs and to investigate their susceptibility to conventional antibiotics, and their public health implications. The results obtain in addition to providing baseline epidemiological data on diseases transmitted from domestic dogs to human, might guide relevant agencies on proactive measures necessary to reducing the trend of zoonotic infectious diseases in our communities.
MATERIALS AND METHODS
Sample Area
Samples were collected from different subunits of Eleme Local Government Area, Rivers State in Southern Nigeria. These subunits are Ebubu, Ekporo, Eteo, Onne, Agbonchia, Akpazo, Alesa, Aleto and Ogale. It lies in greater Port Harcourt metropolis with headquarters in Ogale covering 138 km2 and located in Niger Delta region [20].
Figure 1: Map of the study area, Eleme Local Government Area [20].
Determination of Sample Size
The study sample size was determined using the formula, Sample size (N) = Z2pq/d2. Where, Z is the statistics corresponding to level of 95% confidence level of 1.96. P = expected prevalence in percentage (from similar study) = 0.0333 [21]. d = level of significance (allowable error) of 5%, q = 1-p, 5% = 0.05, q = 1-p = 1 – 0.0333 = 0.967. 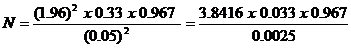 = 49.0. Therefore, a total of 50 samples were collected for the study.
= 49.0. Therefore, a total of 50 samples were collected for the study.
Sample Collection
Nasal and buccal samples were collected aseptically with the aid of swab sticks and transported immediately to the laboratory in a park for microbiological processing such as culture technique. A total of 50 samples were collected from domestic dogs in different location and were cultured on Nutrient, MacConkey, Chocolate and Blood agar accordingly. Spread plate technique was done to achieve the total heterotrophic plate count. All standard procedure for culture were followed and incubated to 24 hours at 37°C to produce discrete colonies and isolated.
Identification of Pathogens Isolated
Gram staining and biochemical tests such as coagulase, citrate, indole, oxidase, urease, and other carbohydrate tests were explore appropriately to identify and classify the pathogens isolated as described by Cheesbrough [22].
Antimicrobial Susceptibility Assay
Kirby-Bauer method (disc method) was explored as described by Cheesbrough [22]. Pure culture plates of isolated organism were selected and colony aseptically emulsified in a sterile saline solution and was mixed thoroughly to ensure that no solid material from the colony is visible in the saline solution. It was repeated until turbidity of the saline solution virtually matched that of the standard turbidity. A standard wire loop used to collect the organism inside the tube and streaked completely on Mueller-Hinton agar (MHA) plate. It was allowed for 5 minutes to dry before antibiotic discs were placed on the surface of the agar using sterile forceps. The disc was gently pressed onto the surface of the agar using flamed sterilized forceps. The inoculated plate was gently inverted and incubated for 24 hours at a temperature of 370C. After incubation, a metric ruler was used to measure the diameter of the zone of inhibition for the antibiotics used. The measurement obtained from the individual antibiotic to the standard table determined whether the tested bacterial species is sensitive or resistance to the tested antibiotic appropriately. The results were categorized based on their standard range and grouped into sensitive, intermediate and non-sensitive.
Statistical Analysis
Data obtained were collated using Microsoft excel and transferred into Statistical Package for Social Science (SPSS) version 21 for analysis. The parameters include mean, standard deviation percentage prevalence, ANOVA and correlation. The significance was observed at probability level of 0.05. Results were presented in figures, tables and charts respectively.
RESULTS
The results obtained from the isolation and antibiogram profile of bacteriological pathogens that are associated with domestic dogs revealed the presence of Gram-negative rod (GNR) bacteria, gram positive rod (GPR) bacteria, gram positive cocci (GPC), and microscopic yeast cells. There was no Gram-negative coccus (GNC) bacterium isolated. However, the overall prevalence was found to be 68% as shown in Figure 1. The prevalence of Escherichia coli, Klebsiella spp., Proteus spp., Bacillus spp., Staphylococcus aureus, and Candida albicans were, 21%, 20%, 7%, 2%, 10%, and 8% respectively.
Table 1 shows the prevalence rate of isolates from different swab sites. Bacillus spp. was isolated from the mouth swab with a prevalence of 2%. Candida albicans, Escherichia coli, Klebsiella spp., Proteus spp., and Staphylococcus aureus have prevalence of 5%, 9%, 12%, 3%, and 4% respectively with total of 35% isolated from the mouth sample. For the nasal swab, the prevalence were 3%, 12%, 8%, 4%, and 6% for Candida albicans, Escherichia coli, Klebsiella spp., Proteus spp., and staphylococcus aureus respectively with total prevalence of 33% isolated.
Table 2 shows the chi square distribution of sensitivity profile of Amoxicillin with swab sites. No growth was found on 15(30.0%) of the plates. However, 5(10%) were resistant to amoxicillin and 2(4.0%) were intermediate for mouth swab when compare to the standard, while in 17(34.0%) of nasal swab, no growth was observed, 4(8.0%), 4(8.0%) and 25(50.0%) were resistant, intermediate and sensitive respectively. The total percentage of resistance to amoxicillin was 9(9.0%) and total percentage of sensitivity was 53(53.0%).
Note: GNB=Gram Negative Bacteria; GPB = Gram Positive Bacteria, GNR = Gram Negative Rod; GNC= Gram Negative Cocci; GPR = Gram Positive Rod; GPC= Gram Positive Cocci
Figure 2: Flowchart showing the prevalence and classification of isolated pathogens.
Table 1: Prevalence rates of pathogenic isolates from different swab sites.
|
|
Bacillus spp. |
Candida albicans |
E. coli |
Klebsiella spp. |
Proteus spp. |
Staph. aureus |
Total |
|
Mouth Swab |
2% |
5% |
9% |
12% |
3% |
4% |
35% |
|
Nasal Swab |
0% |
3% |
12% |
8% |
4% |
6% |
33% |
|
Total |
2% |
8% |
21% |
20% |
7% |
10% |
68% |
Table 2: Distribution of sensitivity profile of amoxicillin with swab sites.
|
|
Amoxicillin-Minimum Inhibition Zone (mm) |
Total |
|||||
|
No Growth |
Resistant (≤14mm) |
Intermediate (15 – 19 mm) |
Sensitive (≥20mm) |
||||
|
|
|
|
|
|
|
||
|
Swab Sites |
|
Mouth Swab |
15 (30.0%) |
5 (10.0%) |
2(4.0%) |
28 (56.0%) |
50 (100.0%) |
|
|
Nasal Swab |
17 (34.0%) |
4 (8.0%) |
4 (8.0%) |
25 (50.0%) |
50 (100.0%) |
|
|
|
Total |
32 (32.0%) |
9 (9.0%) |
6 (6.0%) |
53 (53.0%) |
100(100.0%) |
|
Pearson Chi-Square X2=1.073; DF=3; p=0.78; N=100. Likelihood Ratio = 1.086; DF = 3; p=0.78.
Table 3 shows the Chi square distribution of sensitivity profile of cefuroxime with swab sites. From the total number of organisms that were recovered, 66(66.0%) were resistant to cefuroxime whereas only 2(2.0%) were sensitive to cefuroxime in both the buccal and nasal sample.
Table 4 indicates the Chi square distribution of sensitivity profile of gentamycin with swab sites. The total of 8(8.0%) of the isolated organism were resistant to gentamycin, and a total of 57(57.0%) were sensitive to gentamycin, 32(32.0%) did not show growth, 3(3.0%) were intermediate from both the buccal and nasal samples.
Table 5 indicates the Chi square distribution of sensitivity profile of ciprofloxacin with swab sites. Ciprofloxacin was sensitive to 62(62.0%) and resistant to 6(6.0%) of the total organism isolated.
Table 3: Distribution of sensitivity profile of Cefuroxime with swab sites.
|
|
Cefuroxime |
Total |
||||
|
No Growth |
Resistant (≤14mm) |
Sensitive (≥20mm) |
||||
|
Swab Sites |
|
Mouth Swab |
15 (30.0%) |
33 (66.0%) |
2 (4.0%) |
50 (100.0%) |
|
|
Nasal Swab |
17 (34.0%) |
33 (66.0%) |
0 (0.0%) |
50 (100.0%) |
|
|
|
Total |
32 (32.0%) |
66 66.0% |
2 2.0% |
100 (100.0%) |
|
Table 4: Distribution of sensitivity profile of Gentamycin with swab sites.
|
|
MZI Gentamycin |
Total |
||||
|
No Growth |
Resistant (≤14mm) |
Intermediate (15 - 19mm) |
Sensitive (≥20mm) |
|||
|
Swab Sites |
Mout Swab |
15 (30.0%) |
3 (6.0%) |
2 (4.0%) |
30 (60.0%) |
50 (100.0%) |
|
Nasal Swab |
17 (34.0%) |
5 (10.0%) |
1 (2.0%) |
27 (54.0%) |
50 (100.0%) |
|
|
|
Total |
32 (32.0%) |
8 (8.0%) |
3(3.0%) |
57 (57.0%) |
100 (100.0%) |
Table 5: Distribution of sensitivity profile of Ciprofloxacin with swab sites.
|
|
MZI Ciprofloxacin |
Total |
||||
|
No Growth |
Resistant (≤14mm) |
Sensitive (≥20mm) |
||||
|
Swab Site |
|
Mouth Swab |
15 (30.0%) |
4 (8.0%) |
31 (62.0%) |
50 (100.0%) |
|
|
Nasal Swab |
17 (34.0%) |
2 (4.0%) |
31 (62.0%) |
50 (100.0%) |
|
|
|
Total |
32 (32.0%) |
6 (6.0%) |
62 (62.0%) |
100 (100.0%) |
|
Pearson Chi-Square X2=0.792; DF=2; p=0.67; N=100. Likelihood Ratio = 0.805; DF = 2; p=0.67
Plate 1: Showing antibiotic susceptibility assay of isolates of the study.
Plate 2: Media showing growths of microorganisms and biochemical tests.
DISCUSSION
Droplets from both oral and nasal cavities of domestic dogs in our surrounding may constitute an important source of potential pathogenic microorganisms that would contaminate both dog owners and the community ecosystem at large, hence making the environment unsafe for human habitation. Report of earlier study on dogs and associated pathogens conducted by Kasempimolporn et al. (2003) revealed that varying infections can be acquired from dogs with their various causative microbial agents in our localities. The researcher in their study isolated and identified from oral samples extracted from dogs different bacteria such as Citrobacter freundii, Enterobacter cloacae, Acinetobacter calcoaceticus, and Pasteurella species. However, pathogens isolated from samples from dogs in this study were Bacillus spp., Candida albicans, Escherichia coli, Klebsiella spp., Proteus spp., and Staphylococcus aureus, which are mostly pathogens of public health significance. The generic and specific differences in the isolated pathogens as observed by the researcher of the present and previous study could be due to differences in study settings and geographical location, population investigated or differences in environmental factors. Nonetheless, Campylobacter spp., Salmonella spp., Brucella spp., S. aureus and E. coli have been reported widely by scholars as the most common pathogens found in dogs [8,17,24,25]. Interestingly, this study corroborates the findings of these earlier studies and revealed the presence of Staphylococcus aureus, Escherichia coli, Bacillus spp., Klebsiella spp., Proteus spp., and Candida albicans with various prevalence rates of 10%, 21%, 2%, 20%, 7%, and 8% respectively. In a related study, Luca and colleagues found that Staphylococcus aureus and Escherichia coli were the only similar pathogens recovered from the same site. This observation is consistent with the report of this study, which shows that domestic dogs harbour pathogenic bacteria of clinical relevance. Moreover, Klebsiella spp., Bacillus spp., and Proteus spp. were also isolated. Some of these zoonotic pathogens can cause significant public health problem. Any disease or infection that can be naturally transmitted from vertebrate animals to humans is termed a zoonosis. Currently over 200 types of zoonoses are known [26], which included dog-associated zoonoses. Research has shown that some diseases begin as zoonoses but their causative agents later mutate into human-only strains, which may trigger disruptive impact on public health. Bacillus spp., is a pathogen frequently involved in food poisoning through production of potent toxin and anthrax in domestic animals and humans. Klebsiella spp. causes pneumonia and Proteus spp. is also the cause of urinary tract infection in human. Transmission of pathogens may occur through dog bite and indirectly, through trap of wild animals by dogs harbouring bacteria and are consumed especially, when they are not properly cooked.
Nonetheless, Laura et al. [27] recently reported that home dogs are carrier of antibiotics resistant bacteria strains. Some strains of bacteria pathogens isolated from home dogs were resistant to tetracycline (70%), streptomycin (46%), ampicillin (44%) and nalidixic acid (38.3%). Similarly, microbial resistance of pathogens isolated to four antibiotics such as amoxicillin, cefuroxime, gentamycin, and ciprofloxacin were observed in this study in varying percentages, which corroborates the report of Karl and Colleagues [28] on antimicrobial resistance in bacteria cultivated from samples collected from dogs.
Furthermore, in another related study carried out by Anyanwu et al. [29], the antibiotics susceptibility pattern of potential agents that could be used to treat pathogenic isolates from dogs were highlighted. In their report, E. coli isolated was highly susceptible to gentamycin with 0% resistant making it more potent against the pathogen, followed by amoxicillin with 9.1% resistant while ciprofloxacin was 54.5% resistant against the pathogen. In this study the researcher observed that gentamycin was 57% sensitive, ciprofloxacin was 62% sensitive, amoxicillin was 53% sensitive, and Cefuroxime was 22% sensitive. This report is consistent with similar work done by Anyanwu and others as earlier reported. However, the slight difference in their reported sensitivity patterns could be attributed to variation of resistant genes in the isolated pathogens and the presence of plasmid in their cells. It could also be linked to excessive abuse of antibiotics, especially in developing countries where some citizens indulge in the habit of combining orthodox medicine and native herbs for the treatment of illness. The public health impact of such a high-risk practice may be enormous; it could lead to increase in the burden of antibiotic-resistant strains of pathogens in communities with weak health facilities, thereby making the residents of such communities vulnerable. Furthermore, World Health Organization asserted recently that the use of antibiotics in animals raised for food is widespread and increases the potential for drug-resistant strains of zoonotic pathogens capable of spreading quickly in animal and human populations [26].
CONCLUSION
Pathogenic bacteria are associated with oral and nasal droplets of domestic dogs. These dogs are always in companion with human in day-to-day activities as pet and for other purposes. Notably some disease-causing bacteria isolated from dogs have developed antibiotic resistance which suggests their potential public health concern especially in developing setting such as Nigeria where high-risk practice such as incessant abuse of antibiotics could further exacerbate the burden of antibiotic-resistant pathogenic strains in our communities.
CONFLICT OF INTEREST
None among authors
FUNDING
No fund was received from any agency for the study.
ACKNOWLEDGEMENT
We are grateful to the dog owners in the study area that helped us during the sample collection from the dogs. We are also grateful to some veterinary clinicians that helped us to collect some samples in their various clinics.
REFERENCES
Ashly M. (2021). Pet Pathogens: What dog owners need to know.
Chelsea FM. (2023). The 11 global pet ownership statistics.
Katagiri S, Oliveira-Sequeira T. (2008). Prevalence of dog intestinal parasites and risk perception of zoonotic infection by dog owners in São Paulo State, Brazil. Zoonoses and Public Health. 55(8-10):406–413.
Ghasemzadeh I, Namazi SH. (2015). Review of bacterial and viral zoonotic infections transmitted by dogs. Journal of Medicine and Life. 8(4):1-5.
Rist CL, Arriola CS, Rubin C. (2014). Prioritizing zoonoses: A proposed one health tool for collaborative decision-making. PLoS One. 9(10):e109986.
Grace D, Mutua F, Ochungo P, Kruska R, Jones K, Brierley L, et al. (2012). Mapping of poverty and likely zoonoses hotspots.Zoonoses Project 4, Report to the UK Department for International Development, Nairobi, Kenya: ILRI.
Jones BA, Grace D, Kock R, Alonso S. (2013). Zoonosis emergence linked to agricultural intensification and environmental change. Proceedings of the National Academy of Sciences of the United States of America. 110(21):8399-8404.
Zahra M. (2007).Why you should never let your dog lick your face - as it could be life- threatening.
Day MJ. (2011). Sero-prevalence of vector-borne infection in dogs in Hungary. J Med Life. 4(1):49.
Chomel B. (2014). Emerging and re-emerging zoonosis of dogs and cats. Animals J. 4:434-445.
Mani I, Maguire JH. (2009). Small animal zoonosis and immunocompromised pet owner. J Top Companion Animal Med. 24(4):164-174.
Center for Disease Control. (2018).Public health in action.
Winslow CA. (1920). The untitled field of public health and modern medicine. J Public Health. 2(1):183-191.
Ibrahim MA, Gihan KA, Aboelhadid SM, Abdel-Rahim MM. (2016). Role of pet dogs in transmitting zoonotic intestinal parasites in Egypt. Asian J Animal Vet Adv. 11(6):341-349.
Tracey SM. (2002). Diagnosis and control of zoonotic infections.
Gunaratnam P, Massey PD, Eastwood K., Durrhein D, Graves S, Coote D, et al.(2014). Diagnosis and management of zoonoses - a tool for general practice. National Institute of Health. 43(3):124-128.
Marks SL. (2003). Bacterial gastroenteritis in dogs & cats proceedings of the 28th World Congress of the World Small Animal Veterinary Association.
Veronica OA, Asabe AD, Jarlath UU. (2014). Assessment of knowledge, attitude and practices of dog owners to canine rabies in Wukari Metropolis, Taraba State. Nigeria. Global J Health Sci. 6:226-240.
Leslie EO, Jarlath UU, Asabe AD. (2014). Assessment of risk of possible exposure to rabies among processors and consumers of dog meat in Zaria and Kafanchan, Kaduna State, Nigeria. Global J Health Sci. 61:142-153.
Oyebade BA, Buochuama A. (2016). Tree crown model for a multi-species swamp forest plantation in Nigeria.
Reithinger R, Canales EJ, Llanos-cuentas A, Davies CR. (2003). Domestic dog ownership: A risk factor for human infection with leishmania (viannia) species. Trans R Soc Trop Med Hyg. 97(2):141-145.
Cheesbrough M. (2006). District laboratory practice in tropical countries. 2nd Ed. Cambridge University Press.
Kasempimolporn S, Benjavongkulchai M, Saengseesom W, Sitprija V. (2003). Oral bacterial flora of dogs with and without rabies: A preliminary study in Thailand. J MedAssociation of Thailand. 86(12):1162-1166.
Earnest W. (2018). Campylobacter infection in dogs.
Scott W. (2008). E. coli & dogs.
World Health Organization. (2020). Fact sheet on zoonoses.
Laura M, Daniel B, Lihuel GC, Andrea B, Manuel C, Nora M. (2021). Pet and stray dogs as reservoirs of antimicrobial-resistant Escherichia coli. Int J Microbiol. 2021:6664557.
Karl P, Kristina P, Helene J, Kai F, Vibeke FJ, Ole EH. (2007). Occurrence of antimicrobial resistance in bacteria from diagnostic samples from dogs. J Antimicrob Chemother. 60(4):775–781.
Anyanwu MU, Ugwu IC, Ezekwelu MO, Okafor ON. (2017). Prevalence and antibiogram of generic extended spectrum of beta-lactam resistant enterobacteria in healthy dogs. J Notulae Scientia Biologicae. 9(1):22-33.
.PNG)




